Decomposing heterogeneous dynamical systems with graph neural networks
This site shows the code to generate some figures from the “Decomposing heterogeneous dynamical systems with graph neural networks” paper.
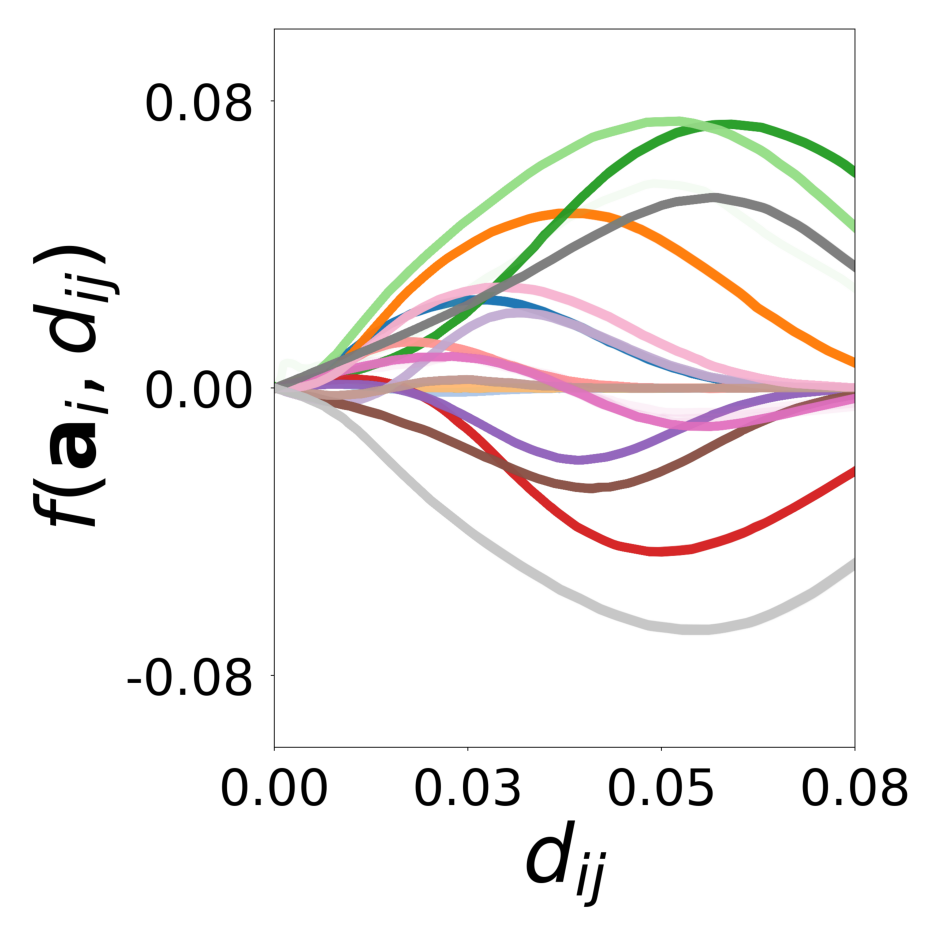
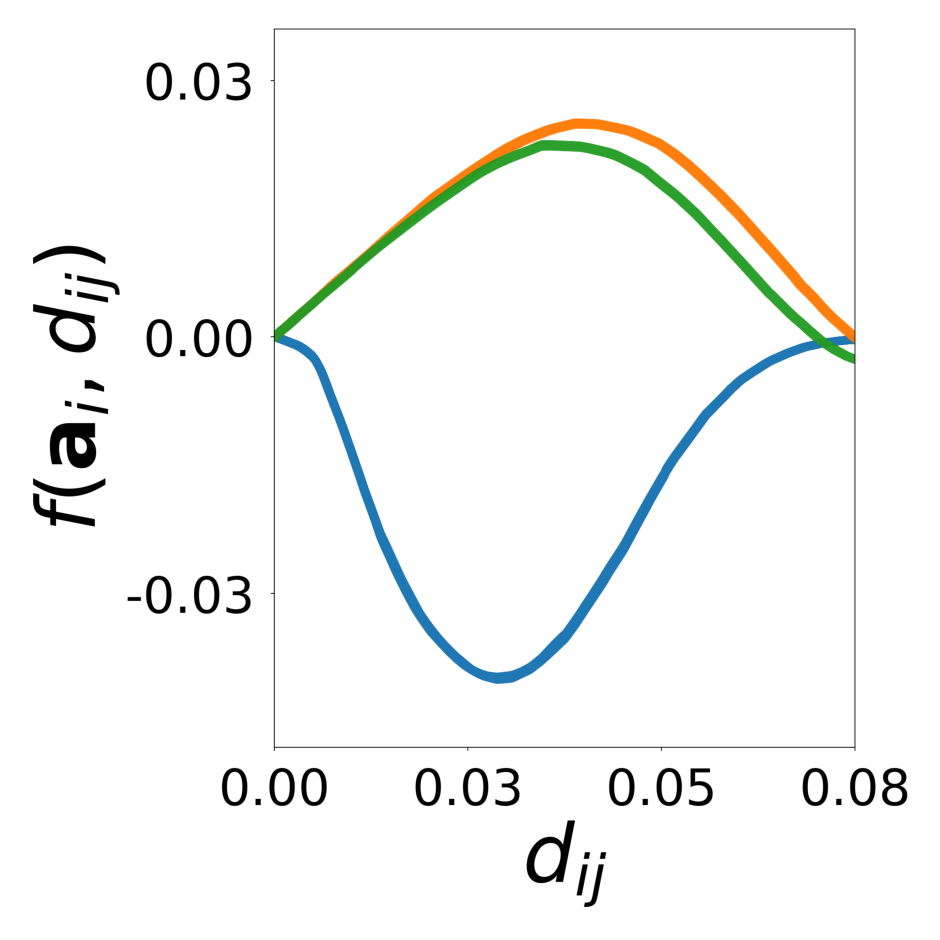
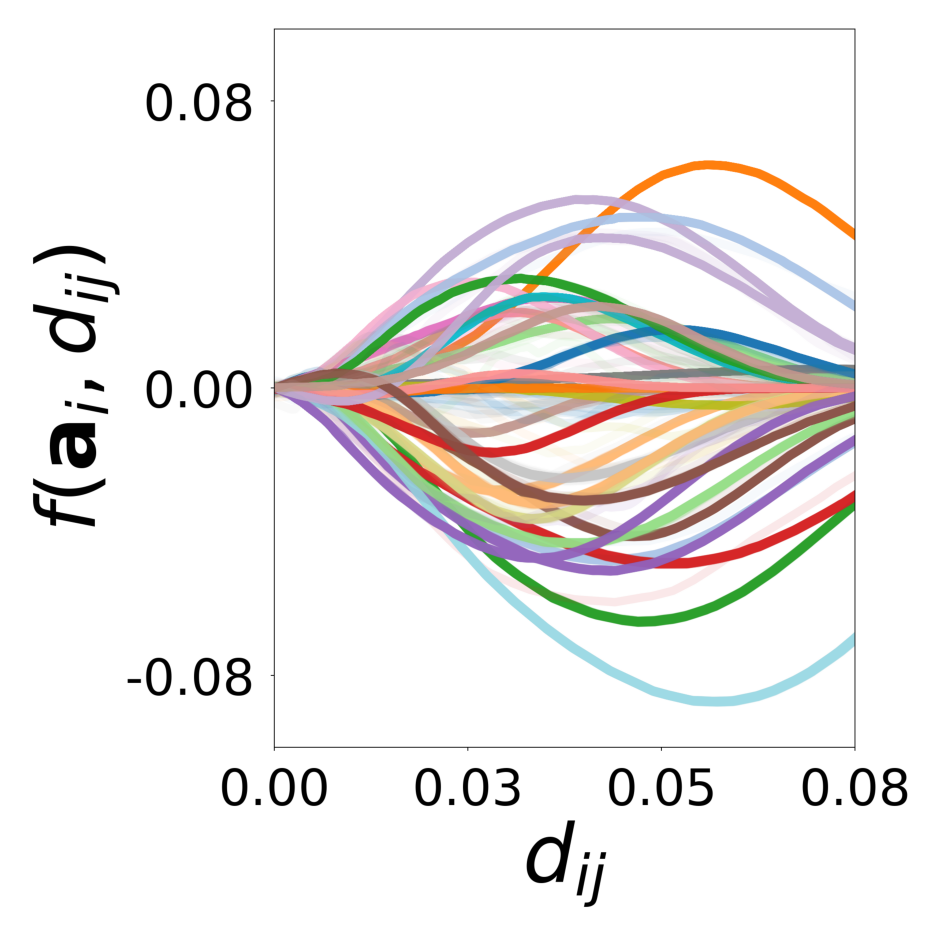
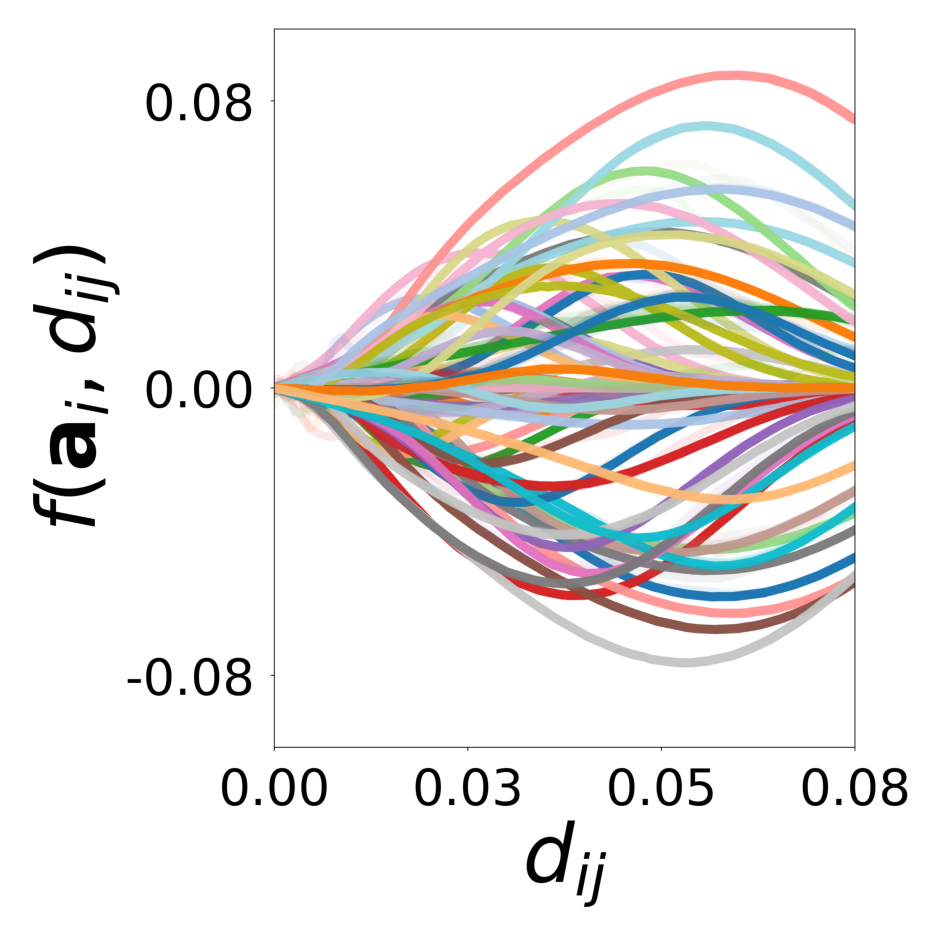
We showed with a diverse set of simulations that message passing GNNs that jointly learn interaction, update functions and latent node properties are a flexible tool to predict, decompose, and eventually understand complex dynamical systems. With the currently available PyTorch Geometric software libraries, it is straightforward to implement a differentiable architecture and loss that encode useful assumptions about the structure of the complex system such as local connectivity rules or the location of learnable and known functions and their inputs. In particular, a well designed GNN can learn a low-dimensional embedding of complex latent properties required to parameterize heterogeneous particle-particle interactions. The learned embeddings can be used to reveal the structure of latent properties underlying the complex dynamics and to infer the corresponding parameters. It is possible to dissect the dynamical system and conduct virtual experiments with arbitrary compositions of particles and interactions. This ability will be particularly important for understanding the behavior of heterogeneous dynamical systems in biology that can only be observed in their natural, mixed configurations. Examples include bacterial community organization, embryonic development, neural networks, and the social interactions of animal communities, which cannot be studied in isolation.