config_file = 'boids_16_256'
config = ParticleGraphConfig.from_yaml(f'./config/{config_file}.yaml')
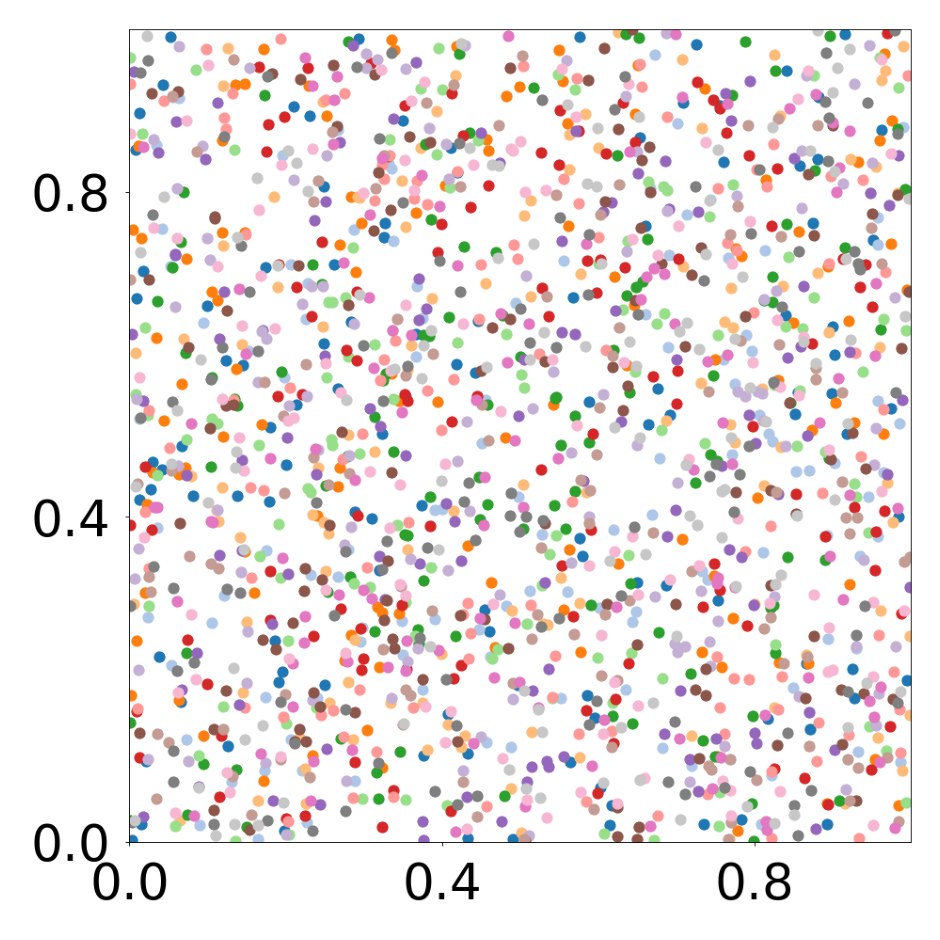
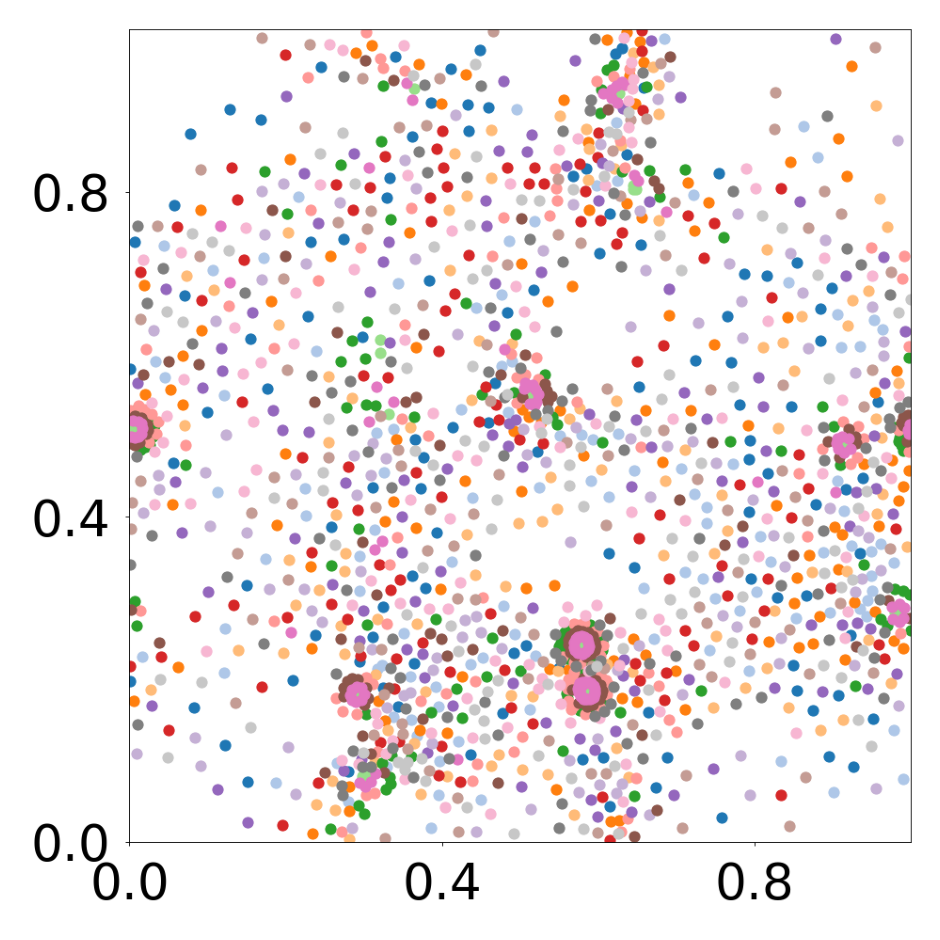
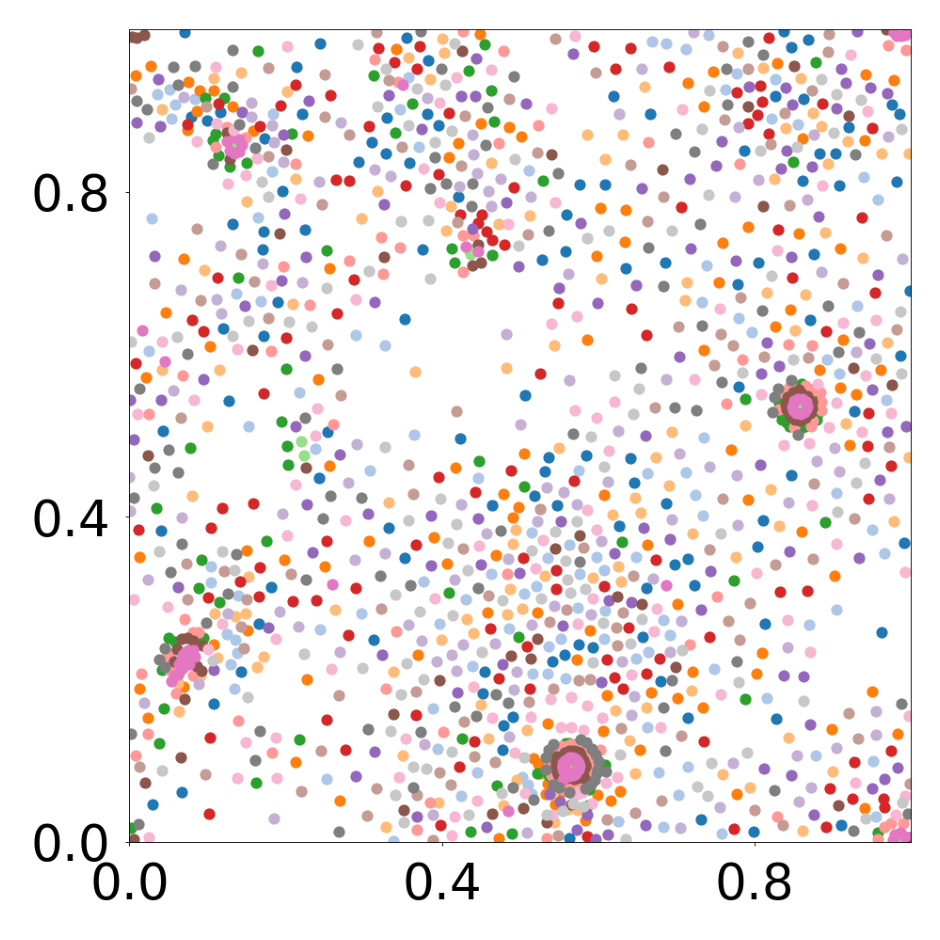
device = set_device("auto")Boids system with 16 different particle types
Particles
Simulation
This script creates the fourth column of paper’s Figure 2. Simulation of boids (https://en.wikipedia.org/wiki/Boids), 1792 particles, 16 types.
First, we load the configuration file and set the device.
The following model is used to simulate the boids system with PyTorch Geometric.
class BoidsModel(pyg.nn.MessagePassing):
"""Interaction Network as proposed in this paper:
https://proceedings.neurips.cc/paper/2016/hash/3147da8ab4a0437c15ef51a5cc7f2dc4-Abstract.html"""
"""
Compute the acceleration of Boids as a function of their relative positions and relative positions.
The interaction function is defined by three parameters p = (p1, p2, p3)
Inputs
----------
data : a torch_geometric.data object
Returns
-------
pred : float
the acceleration of the Boids (dimension 2)
"""
def __init__(self, aggr_type=[], p=[], bc_dpos=[], dimension=2):
super(BoidsModel, self).__init__(aggr=aggr_type) # "mean" aggregation.
self.p = p
self.bc_dpos = bc_dpos
self.dimension = dimension
self.a1 = 0.5E-5
self.a2 = 5E-4
self.a3 = 1E-8
self.a4 = 0.5E-5
self.a5 = 1E-8
def forward(self, data=[], has_field=False):
x, edge_index = data.x, data.edge_index
if has_field:
field = x[:,6:7]
else:
field = torch.ones_like(x[:,0:1])
edge_index, _ = pyg_utils.remove_self_loops(edge_index)
particle_type = to_numpy(x[:, 1 + 2*self.dimension])
parameters = self.p[particle_type, :]
d_pos = x[:, self.dimension+1:1 + 2*self.dimension].clone().detach()
dd_pos = self.propagate(edge_index, pos=x[:, 1:self.dimension+1], parameters=parameters, d_pos=d_pos, field=field)
return dd_pos
def message(self, pos_i, pos_j, parameters_i, d_pos_i, d_pos_j, field_j):
distance_squared = torch.sum(self.bc_dpos(pos_j - pos_i) ** 2, axis=1) # distance squared
cohesion = parameters_i[:,0,None] * self.a1 * self.bc_dpos(pos_j - pos_i)
alignment = parameters_i[:,1,None] * self.a2 * self.bc_dpos(d_pos_j - d_pos_i)
separation = - parameters_i[:,2,None] * self.a3 * self.bc_dpos(pos_j - pos_i) / distance_squared[:, None]
return (separation + alignment + cohesion) * field_j
def bc_pos(x):
return torch.remainder(x, 1.0)
def bc_dpos(x):
return torch.remainder(x - 0.5, 1.0) - 0.5The data is generated with the above Pytorch Geometric model. Note two datasets are generated, one for training and one for validation.
p = torch.squeeze(torch.tensor(config.simulation.params))
model = BoidsModel(aggr_type=config.graph_model.aggr_type, p=torch.squeeze(p), bc_dpos=bc_dpos, dimension=config.simulation.dimension)
generate_kwargs = dict(device=device, visualize=True, run_vizualized=0, style='color', alpha=1, erase=True, save=True, step=10)
train_kwargs = dict(device=device, erase=True)
test_kwargs = dict(device=device, visualize=True, style='color', verbose=False, best_model='20', run=0, step=1, save_velocity=True)
data_generate_particles(config, model, bc_pos, bc_dpos, **generate_kwargs)Finally, we generate the figures that are shown in Figure 2. All frames are saved in ‘decomp-gnn/paper_experiments/graphs_data/graphs_boids_16_256/Fig/’.